【 References 】
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
2. Ajani JA, D'Amico TA, Bentrem DJ, et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:855-83.
3. Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet 2017;390:2383-96.
4. Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102.
5. Wang F, Liu DB, Zhao Q, et al. The genomic landscape of small cell carcinoma of the esophagus. Cell Res 2018;28:771-4.
6. Wang S, Xiong Y, Zhang Q, et al. Clinical signifcance and immunogenomic landscape analyses of the immune cell signature based prognostic model for patients with breast cancer. Brief Bioinform 2020;bbaa311.
7. Zeng D, Li M, Zhou R, et al. Tumor Microenvironment Characterization in Gastric Cancer Identifes Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol Res 2019;7:737-50.
8. Zheng Y, Chen Z, Han Y, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun 2020;11:6268.
9. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74.
10. Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717-34.
11. Galluzzi L, Buqué A, Kepp O, et al. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015;28:690-714.
12. Galluzzi L, Senovilla L, Zitvogel L, et al. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov 2012;11:215-33.
13. Fridman WH, Galon J, Pagès F, et al. Prognostic and predictive impact of intra- and peritumoral immune infltrates. Cancer Res 2011;71:5601-5.
14. Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306.
15. Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore Signature: A Prognostic and Predictive Tool in Gastric Cancer. Ann Surg 2018;267:504-13.
16. Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20.
17. Le DT, Durham JN, Smith KN, et al. Mismatch repair defciency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13.
18. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218.
19. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95.
20. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infltrating immune cells across human cancers. Nat Med 2015;21:938-45.
21. Stoll G, Bindea G, Mlecnik B, et al. Meta-analysis of organ-specifc differences in the structure of the immune infltrate in major malignancies. Oncotarget 2015;6:11894-909.
22. Stoll G, Enot D, Mlecnik B, et al. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014;3:e27884.
23. O’Sullivan JA, Bochner BS. Eosinophils and eosinophilassociated diseases: An update. J Allergy Clin Immunol 2018;141:505-17.
24. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889-96.
25. Zheng QX, Wang J, Gu XY, et al. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomed Pharmacother 2021;135:111169.
26. Lin C, Zhang S, Wang Y, et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res 2018;24:486-98.
27. Zhang W, Hong R, Xue L, et al. Piccolo mediates EGFR signaling and acts as a prognostic biomarker in esophageal squamous cell carcinoma. Oncogene 2017;36:3890-902.
28. Zhang H, Song J, Dong J, et al. Tumor Microenvironment Analysis Identifed Subtypes Associated With the Prognosis and the Tumor Response to Immunotherapy in Bladder Cancer. Front Genet 2021;12:551605.
29. Lee HK, Kwon MJ, Ra YJ, et al. Signifcance of druggable targets (PD-L1, KRAS, BRAF, PIK3CA, MSI, and HPV) on curatively resected esophageal squamous cell carcinoma. Diagn Pathol 2020;15:126.
30. Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A 2010;107:4275-80.
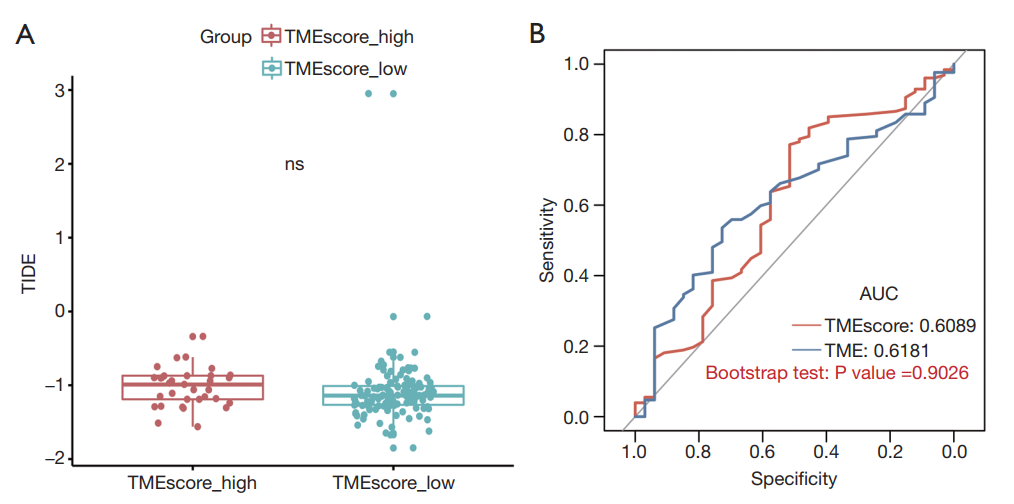
31. TIDE: Tumor Immune Dysfunction and Exclusion. Available online: http://tide.dfci.harvard.edu/
32. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6.
33. Galon J, Pagès F, Marincola FM, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med 2012;10:205.
34. Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classifcation of cancer. J Transl Med 2012;10:1.
35. Reichman H, Karo-Atar D, Munitz A. Emerging Roles for Eosinophils in the Tumor Microenvironment. Trends Cancer 2016;2:664-75.
36. Collins PD, Marleau S, Griffths-Johnson DA, et al. Cooperation between interleukin-5 and the chemokine eotaxin to induce eosinophil accumulation in vivo. J Exp Med 1995;182:1169-74.
37. Rothenberg ME. Eosinophilia. N Engl J Med 1998;338:1592-600.
38. Sanderson CJ. Interleukin-5, eosinophils, and disease. Blood 1992;79:3101-9.
39. Panganiban RP, Pinkerton MH, Maru SY, et al. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol 2012;1:154-65.
40. Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget 2017;8:624-43.
41. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50.
42. Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017;171:934-949.e16.
43. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71.
44. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8.